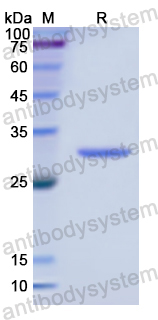
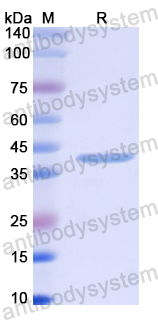
Catalog No.
YHD97401
Expression system
E. coli
Species
Homo sapiens (Human)
Protein length
Met1-Gln242
Predicted molecular weight
29.99 kDa
Nature
Recombinant
Endotoxin level
Please contact with the lab for this information.
Purity
>90% as determined by SDS-PAGE.
Accession
P31944
Applications
ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Form
Lyophilized
Storage buffer
Lyophilized from a solution in PBS pH 7.4, 0.02% NLS, 1mM EDTA, 4% Trehalose, 1% Mannitol.
Reconstitution
Reconstitute in sterile water for a stock solution. A copy of datasheet will be provided with the products, please refer to it for details.
Shipping
In general, proteins are provided as lyophilized powder/frozen liquid. They are shipped out with dry ice/blue ice unless customers require otherwise.
Stability and Storage
Use a manual defrost freezer and avoid repeated freeze thaw cycles. Store at 2 to 8°C for frequent use. Store at -20 to -80°C for twelve months from the date of receipt.
Alternative Names
CASP14, CASP-14, Caspase-14
Exhaled breath protein composition in patients hospitalised during the first wave of COVID-19., PMID:40341493
LightRoseTTA: High-Efficient and Accurate Protein Structure Prediction Using a Light-Weight Deep Graph Model., PMID:40134034
Tissue-specific and functional loci analysis of CASP14 gene in the sheep horn., PMID:39570959
Mechanism of ginsenoside Rb3 against OGD/R damage based on metabonomic and PCR array analyses., PMID:39420925
AI-Driven Deep Learning Techniques in Protein Structure Prediction., PMID:39125995
Comparative genomics of sirenians reveals evolution of filaggrin and caspase-14 upon adaptation of the epidermis to aquatic life., PMID:38653760
MFTrans: A multi-feature transformer network for protein secondary structure prediction., PMID:38599417
Integration Analysis of Hair Follicle Transcriptome and Proteome Reveals the Mechanisms Regulating Wool Fiber Diameter in Angora Rabbits., PMID:38542234
Atom-ProteinQA: Atom-level protein model quality assessment through fine-grained joint learning., PMID:38537495
BDM: An Assessment Metric for Protein Complex Structure Models Based on Distance Difference Matrix., PMID:38536590
Novel Prognostic Model Construction of Tongue Squamous Cell Carcinoma Based on Apigenin-Associated Genes., PMID:38420803
Recent Progress of Protein Tertiary Structure Prediction., PMID:38398585
PSSP-MFFNet: A Multifeature Fusion Network for Protein Secondary Structure Prediction., PMID:38343972
Protein language models meet reduced amino acid alphabets., PMID:38310333
Solving protein structures by combining structure prediction, molecular replacement and direct-methods-aided model completion., PMID:38214490
Assessing protein model quality based on deep graph coupled networks using protein language model., PMID:38018909
H-Packer: Holographic Rotationally Equivariant Convolutional Neural Network for Protein Side-Chain Packing., PMID:38013891
Role of environmental specificity in CASP results., PMID:37950210
Critical assessment of methods of protein structure prediction (CASP)-Round XV., PMID:37920879
The impact of AI-based modeling on the accuracy of protein assembly prediction: Insights from CASP15., PMID:37861057
Evaluation of the Ability of AlphaFold to Predict the Three-Dimensional Structures of Antibodies and Epitopes., PMID:37782047
Tertiary structure assessment at CASP15., PMID:37746927
Improving AlphaFold2-based protein tertiary structure prediction with MULTICOM in CASP15., PMID:37679431
Predicted models and CCP4., PMID:37594303
The Impact of AI-Based Modeling on the Accuracy of Protein Assembly Prediction: Insights from CASP15., PMID:37503072
More than just pattern recognition: Prediction of uncommon protein structure features by AI methods., PMID:37399411
iQDeep: an integrated web server for protein scoring using multiscale deep learning models., PMID:37356909
GraphGPSM: a global scoring model for protein structure using graph neural networks., PMID:37317619
An end-to-end deep learning method for protein side-chain packing and inverse folding., PMID:37253017
Protein model quality assessment using rotation-equivariant transformations on point clouds., PMID:37158708
Atomic protein structure refinement using all-atom graph representations and SE(3)-equivariant graph transformer., PMID:37144951
Progress at protein structure prediction, as seen in CASP15., PMID:37060758
Ensemble deep learning models for protein secondary structure prediction using bidirectional temporal convolution and bidirectional long short-term memory., PMID:36860882
High-accuracy protein model quality assessment using attention graph neural networks., PMID:36736352
DLBLS_SS: protein secondary structure prediction using deep learning and broad learning system., PMID:36505696
An integrated protein structure fitness scoring approach for identifying native-like model structures., PMID:36467582
Improved model quality assessment using sequence and structural information by enhanced deep neural networks., PMID:36460624
Convolutional ProteinUnetLM competitive with long short-term memory-based protein secondary structure predictors., PMID:36448315
Gallic acid has an inhibitory effect on skin squamous cell carcinoma and acts on the heat shock protein HSP90AB1., PMID:36375658
Improving protein structure prediction using templates and sequence embedding., PMID:36355462
Improved Protein Real-Valued Distance Prediction Using Deep Residual Dense Network (DRDN)., PMID:36008645
Estimation of model accuracy by a unique set of features and tree-based regressor., PMID:35982086
Studying protein-protein interaction through side-chain modeling method OPUS-Mut., PMID:35959990
Training undergraduate research assistants with an outcome-oriented and skill-based mentoring strategy., PMID:35916219
The integration of AlphaFold-predicted and crystal structures of human trans-3-hydroxy-l-proline dehydratase reveals a regulatory catalytic mechanism., PMID:35891782
Multi-head attention-based U-Nets for predicting protein domain boundaries using 1D sequence features and 2D distance maps., PMID:35854211
Predicting residue-specific qualities of individual protein models using residual neural networks and graph neural networks., PMID:35842895
Collective Variable for Metadynamics Derived From AlphaFold Output., PMID:35769910
Protein structure prediction improves the quality of amino-acid sequence alignment., PMID:35754316