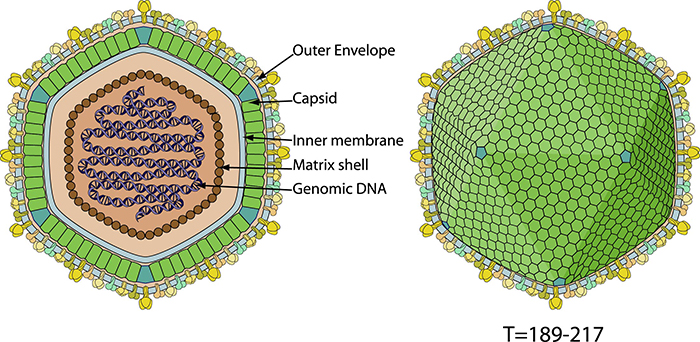
The extracellular ASFV particle structure (Swiss institute of Bioinformatics)
African swine fever virus (ASFV)
African swine fever (ASF) is a highly contagious, widespread hemorrhagic swine infectious disease caused by African swine fever virus (ASFV). The World Organization for Animal Health (OIE) lists it as a statutory reportable animal disease. In China, it is listed as a first-class animal disease and one of the key exotic animal diseases to guard against.
ASFV is an enveloped double-stranded DNA virus, the only member of the genus Asfivirus of the African swine fever-associated virus family (Asfarviridae), and the only DNA arbovirus discovered so far. The extracellular virion is about 200nm in diameter and consists of a concentric multilayer icosahedral symmetrical structure formed by Internal core, matrix shell, Inner membrane, capsid, and outer envelope.
The ASFV genome is about 170-190 kbp in length and encodes more than 150 polypeptides, of which 30-50 are structural proteins that constitute viral particles. In addition to structural proteins, various enzymes related to DNA transcription and replication, protein modification, and various proteins related to virus-host interactions are also encoded. Studies have shown that more than 50 proteins are packaged into virions and play a role in viral infection, such as p54, p30, CD2v, pp220, pp62, p72, etc. pp220 and pp62 are the polyprotein precursors of ASFV; p54 and p30 are crucial antigenic proteins; p72 is mainly involved in the composition of the capsid, and plays an important role in the late expression of virus infection and the formation of the viral capsid ; CD2v is a glycoprotein that plays a prominent role in the pathogenesis of African swine fever and has additional roles in virus tissue tropism and immune escape to the host; p10, p12, p14.5 and p17 proteins are involved in virion adsorption and transfer.
The viral proteins identified in the ASFV proteome can be divided into the following functional categories: involved in viral structure formation and morphological maintenance, accounting for 24% of the total protein; involved in viral transcription and RNA modification, accounting for 19% of the total protein; maintaining genome Integrity, accounting for 6% of the total protein; mediating virus entry, accounting for 4% of the total protein; assisting the virus to escape host defense, accounting for 3% of the total protein; proteins that have been studied but whose functions are unknown, accounting for 10% of total protein; unknown proteins remained the most abundant category, accounting for 34% of total protein. Studies have shown that most of these detected proteins are located in the Nucleoid layer, mainly including proteins pA104R and pK78R that bind to the genome, transcription factors involved in mRNA transcription and modification, viral dUTPase, and three enzymes involved in DNA repair. The IAP homolog pA224L. Since the inner lipid membrane should contain the proteins required to transform and stabilize the endoplasmic reticulum-derived membrane into the viral membrane, while supporting the multiple interactions required for the outer capsid and inner nucleocapsid, mediate the fusion mechanism after viral entry into the host cell , therefore Innercapsid has at least seven known viral membrane proteins: pD117L, pE183L, p12pO61R, pKP177L, pH108R, pE199L and pE248R. Only one viral membrane protein, CD2v, is located in the Outer envelope layer. The Outer capsid layer is mainly composed of the capsid protein p72 and the minor capsid protein p49, and also contains the protein pE120R that mediates intracellular viral trafficking.
In recent years, with the deepening of the research on the structure and function of the protein encoded by ASFV, the structure and infection process of ASFV have gradually become clear. However, due to the large number of ASFV genomes and the complex immune escape mechanism, the pathogenic mechanism of the virus remains unclear. More research is needed to develop an effective vaccine against ASFV.
Polyprotein pp220(Q08358)
Function
Essential for the core assembly. Its myristoyl moiety may function as a membrane-anchoring signal to bind the developing core shell to the inner viral envelope.
Polyprotein pp62(Q65179)
Function
Essential for the correct assembly and maturation of the core of the virion.
Inner membrane protein p54(Q65194)
Function
Inner envelope protein involved, through its interaction with host dynein, in the intracellular microtubule-dependent transport of viral capsid toward viral factories.
Seems to induce caspase-3 activation and apoptosis. Plays a role in virion morphogenesis by recruiting and transforming the host ER membranes into the precursors of the viral envelope. Involved in virus attachment to the host cell.
Phosphoprotein p30(P34204)
Function
Modifies the subcellular distribution of heterogeneous nuclear ribonucleoprotein K (HNRNPK) and may contribute to modulate HNRNPK functions related to processing and export of mRNAs during ASFV infection. Necessary for virus internalization.
CD2 homolog(Q89501)
Function
May play an immunosuppressive role by inhibiting lymphocyte proliferation and subsequently facilitating viral replication and generalization of infection (By similarity). Responsible for viral hemadsorption, which may help viral spread. Increases virus replication in the tick vector at the step of virus uptake or replication in the tick gut. May play a role in the host Golgi reorganization to yield viral factories. May play a role in host cell penetration.
Hexon protein p72(P22776)
Function
Capsid protein that self-assembles to form the pseudo-hexameric capsomers of the icosahedral capsid. The capsid is constructed of 2760 pseudo-hexameric capsomers and 12 pentameric capsomers, with a T=277 symmetry, about 200 nm in diameter.
The capsid encapsulates the DNA-containing nucleoid, the core shell and the inner membrane. Plays an essential role in virion assembly. Involved in virus attachment to the host cell.
Reference:
1.African Swine Fever Virus: A Review. PMID: 28489063
2.African Swine Fever Virus Biology and Vaccine Approaches. PMID: 29551143
3.African swine fever virus replication and genomics. PMID: 23142553
4.African swine fever. PMID: 19967933
5.African Swine Fever Virus MGF-505-7R Negatively Regulates 6.cGAS-STING-Mediated Signaling Pathway. PMID: 33712518
7.Architecture of African swine fever virus and implications for viral assembly. PMID: 31624094
8.Molecular Characterization of African Swine Fever Virus, China, 2018. PMID: 9.30141772
10.Cryo-EM Structure of the African Swine Fever Virus. PMID: 31787524
11.A Proteomic Atlas of the African Swine Fever Virus Particle. PMID: 30185597
12.African Swine Fever Virus MGF-110-9L-deficient Mutant Has Attenuated Virulence in Pigs. PMID: 33689140
13.Genetic and antigenic diversity of African swine fever virus. PMID: 31330205
14.Epitope mapping of African swine fever virus (ASFV) structural protein, p54. PMID: 32004574
15.African Swine Fever Virus Protein pE199L Mediates Virus Entry by Enabling 16.Membrane Fusion and Core Penetration. PMID: 32788374
17.High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. PMID: 32056963
18.African Swine Fever Virus as a Difficult Opponent in the Fight for a Vaccine-Current Data. PMID: 34201761
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: UUH61865.1
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: YP_009927243.1
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: YP_009927204.1
Host species: Rabbit
Isotype: IgG
Applications: ELISA, IHC, WB
Accession: YP_009927231.1
Applications: SDS-PAGE, WB, ELISA, Immunogen, Bioactivity testing in progress
Expression system: E. coli
Accession: P23169
Protein length: Phe26-Ala177
Applications: SDS-PAGE, WB, ELISA, Immunogen, Bioactivity testing in progress
Expression system: E. coli
Accession: Q65194
Protein length: Val40-Arg111
Applications: ELISA, Immunogen, SDS-PAGE, WB, Bioactivity testing in progress
Expression system: E. coli
Accession: YP_009927243.1
Protein length: Met1-Lys241